Virtualization of computing environments
Contents
Virtualization of computing environments¶
We all know the problem: we want to run or re-run an analysis but basically nothing works… . Trying to solve installation issues creates more problems than it solves, software dependencies are not compatible, the analyses need a certain OS and chances are high that even if things run, results vary throughout machines. And the “worst part”: your colleagues/collaborators just say the following.

The harsh truth is that computing environments are one of the major aspects one needs to address regarding reproducibility, also in neuroimaging. This refers to the computational architecture one is using, including the respective software stack and versions thereof. But what can be done here? Sending machines around via post? Rather not… However, there’s a process with accompanying resources and tools that is a staple in other research fields since a while but is now also more and more utilized within neuroimaging: virtualization of computing environments. Within this 2 h session of the workshop, we will explore the underlying problems, rationales and basics, as well as provide first hands-on experience.
Content 💡👩🏽🏫¶
In the following you’ll find the objectives and materials for each of the topics we’ll discuss during this session. Specifically, we will get to know virtualization based on a real-world example, i.e. a small python script that used DIPY to perform a set of DTI analyses. The main content and information will be provided as slides but there will also be some scripts. Thus, please check the materials section carefully. This also means will have a split between presenting slides and running things in the terminal.
Objectives 📍¶
Learn about open and reproducible methods and how to apply them using
condaandDocker(orSingularity)Know the differences between
virtualization techniquesFamiliarize yourself with the
virtualization/containerecosystem for scientific workEmpower you with tools and technologies to do
reproducible,scalableandefficientresearch
Materials 📓¶
As mentioned above, we will have a set of different materials for this session, including slides and scripts. The slides include the background information, as well as most of the commands we will run in the terminal during the session. You can find them here or can directly download them:
The scripts entail a python script called fancy_DTI_analyses.py which will be the example on which we will explore virtualization and virtualization_commands.sh which is a bash script that contains all commands we are going to run during the session so that you can easily copy-paste them/have them on file in case you missed something. You can find them in the GitHub repository of this workshop or download them below:
Please make sure to get them on way or the other and place them on your Desktop for easy access. Also, you might want to download the Docker image we are going to build during the session in advance to have it ready to go. You can find it below:
and download it via:
docker pull peerherholz/millennium_falcon:v0.0.1
Questions you should be able to answer based on this lecture 🖥️✍🏽📖¶
What is virtualization and why is it important/helpful?
Virtualization refers to the process of encapsulating computing environments in a way that they can be shared and utilized on different machines. Depending on the virtualization type and problem at hand, it can help a great deal with software/computing management and reproducibility as common issues like installation problems, software dependencies and sustainability can be efficiently tackled.
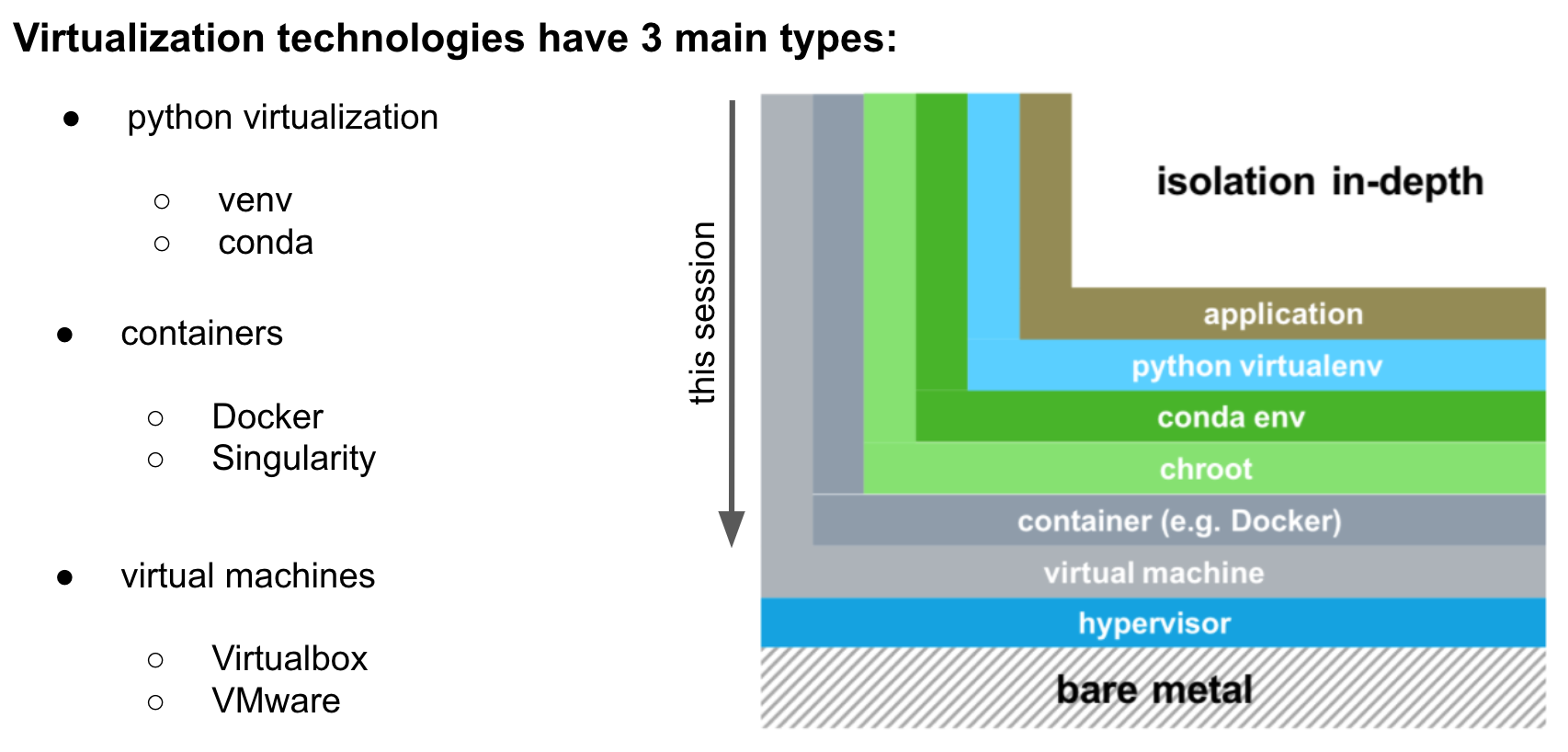
What types of virtualization do exist?
There are three main levels of virtualization as summarized below:
optional reading/further materials¶
There are a lot of fantastic resources out there to further familiarize yourself with virtualization, no matter of dedicated workshops, videos or what have you. Below, we just compiled a small list of other introductory level resources through which you can continue to explore this amazing approach to data management & analyses.

