Overview
Teaching: 300 min Exercises: 40 minQuestions
How do version control systems facilitate reproducibility, and which systems should be used?
Objectives
Become familiar with version control systems for code and data, as well as relevant tools based on them
Learn how to use version control systems to obtain, maintain, and share code and data
Review available third party services and workflows that could be used to help to guarantee reproducibility of results
You can skip this lesson if you can answer these questions:
- How do you keep all your scripts, configuration, notes, and data under version control systems and shareable with your collaborators?
- How do you establish and use continuous integration systems to verify the correctness of reproduced results (where feasible)?
- What exactly did you do in your data analysis project X on date Y?
What is a “version control system”?
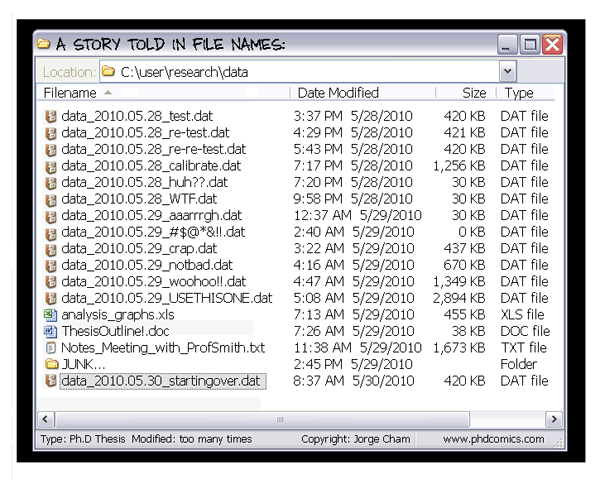
We all probably do some level of version control with our files, documents, and even data – but without a version control system (VCS), we do it in an ad-hoc manner:
In general, a VCS helps you track versions of digital artifacts, such as code (scripts, source files), configuration files, images, documents, and data – both original or derived (e.g., the outcome of an analysis). With proper annotation of changes, a VCS becomes the lab notebook for changing content in the digital world. Since all versions are stored, VCS makes it possible to provide any previous version at a later point in time. You can see how this is critical for reproducing previous results – if your work’s history is stored in a VCS, you just need to get a previous version of your materials to reproduce an earlier analysis. You can also recover a file which you mistakenly removed since a previous version would be contained within your VCS, so no more excuses like “the cat ate my source code”. These features alone make it worthwhile to place any materials you produce and care about under an appropriate VCS.
Besides tracking changes, another main function of a VCS is collaboration. Any modern VCS supports transfer and aggregation of changes to your work among collaborators. Public versioning and collaboration services (such as GitHub) allow you to integrate other online services (such as travis-ci) that can be configured to automatically evaluate any new changes. Integration with such services, which allow data to be automatically reanalyzed and verified for expected results, plays an important role in reproducibility.
In this module we will learn about:
- General use of Git as a VCS to maintain not-so-large files (code, configuration, text, etc.)
- 3rd-party services that integrate with your VCS on public portals (e.g., on GitHub)
- VCS tools for managing data files (git-annex, DataLad)
- Additional VCS-based utilities that can help you to assert greater control over your digital research artifacts and notes
Git
External teaching materials
To gain good general working knowledge of VCSs and Git, please go through the following lessons/tutorials:
- Software Carpentry: Version Control with Git (full: 2:30h, familiarize: 20m) – a thorough lesson of the main Git commands and workflows; please complete the lesson until at least the
Licensingsubmodule, which will be a covered in a separate lecture- Curious Git: A Curious Tale (full: 30m, familiarize: 10m) – useful read if you feel that “Git internals” look like a black box to you; this example guides you through the principles of Git without talking about Git
- (very optional, since this module is Git-based) Software Carpentry: Version Control with Mercurial (full: 4h)
Setting up Git for the first time?
When setting up Git on a new host, we recommend configuring Git so that commits have appropriate author information.
% git config --global user.name "FirstName LastName" % git config --global user.email "ideally@real.email"
Exercise: a basic Git/GitHub workflow
Submit a pull request (PR) suggesting a change to the https://github.com/ReproNim/simple_workflow analysis. You should submit an initial PR with one of the changes, and then improve it with subsequent additional commits, and see how the PR gets automatically updated. Suggested changes for the first commit to initiate a PR:
- a completely dummy change to README.md (0 points)
- fix an actual typo in README.md (10 points)
Then proceed to enact more meaningful change:
- adjust
circle.ymlto run analysis (look for line withrun_demo_workflow.py) on just a single subject (currently-n 2to run on two subjects)
Exercise: exploiting Git history
Goal: determine how the estimate for the Left-Amygdala in AnnArbor_sub16960 subject changed from release 1.0.0 to 1.1.0.
Answer
git diff allows us to see the differences between points in the Git history and to optionally restrict the search to the specific file(s), so the answers to the challenge were
git tagandgit grep:% git diff 1.0.0..1.1.0 -- expected_output/AnnArbor_sub16960/segstats.json ... "Left-Amygdala": [ - 619, - 742.80002951622009 + 608, + 729.60002899169922 ],
Third-party services
As you learned in the Remotes in GitHub section of the Software Carpentry Git course, the GitHub website provides you with public (or private) storage for your Git repositories on the web. The GitHub website also allows third-party websites to interact with your repositories to provide additional services, typically in response to new changes to your repositories. Visit GitHub Marketplace for an overview of the vast collection of such additional services. Some services are free, some are “pay-for-service”. Students can benefit from obtaining a Student Developer Pack to gain free access to some services which otherwise would require a fee.
Continuous integration
A growing number of online services provide continuous integration (CI) services. Although the free tier may not provide sufficient resources to carry out entire analyses on your data, we encourage using CIs. They help verify your code’s correct execution and the reproducibility of your results. CIs can be used to execute unit-tests on simulated data or a subset of the real data. For example, see simple workflow code for a very simple, re-executable neuroimaging publication.
Travis CI
Travis CI was one of the first free continuous integration services integrated with GitHub, and is free for publicly available projects.
External teaching materials
- A quick Travis CI Tutorial for Node.js developers (full: 20m) – a good description of all necessary steps to enable Travis CI for your GitHub project; although geared toward Node.js projects, the same principles apply to other platforms/languages
- Shablona - A template for small scientific python projects (review: 5m, optional) – a template for scientific Python projects; review its
.travis.ymlfor an example of a typical setup for a Python-based project- Travis CI Documentation (familiarize: 10m, canonical reference) – documentation for Travis CI; review sections relevant to your language/platform
CircleCI
External teaching materials
- CircleCI 1.0 Documentation (familiarize: 10m, canonical reference) – documentation for CircleCI; review sections relevant to your language/platform
External review materials
- Continuous Integration in the Cloud: Comparing Travis, Circle and Codeship (review: 10m) – having acquainted yourself with the basics of two CIs, review the differences
- Side-by-side comparison of CI services: review 5m
Exercise: adjust
simple_workflowAdjust
simple_workflowto execute sample analysis on another subject.
git-annex
git-annex is a tool that allows a user to manage data within a Git repository without committing the (large) content of those files directly under git. In a nutshell, git-annex
- moves actual data file(s) under
.git/annex/objectsinto a file typically named according to the checksum of the file’s content, and in its place creates a symbolic link (symlink) pointing to that new location - commits that symlink (not actual data) under git, so a file of any size has the same small footprint within git
- within
git-annexbranch, the location of the data file (on which machine, clone, or web URL) is recorded
Later on, if you have access to the clones of the repository containing
the file, you can easily get it (which will download/copy that file
under .git/annex/objects) or drop it (which will remove that file from
.git/annex/objects).
Since Git doesn’t contain the actual content of large files, but
instead just contains symlinks and information in the git-annex branch, it
becomes possible to
- have very lean Git repositories pointing to arbitrarily large files
- share such repositories on any Git hosting portal (e.g., GitHub);
remember to push the
git-annexbranch as well - very quickly switch (i.e.,
checkout) between different states of the repository because no large files need to be created – just symlinks
Note
Never manually git merge a git-annex branch; git-annex uses a special merge
algorithm to merge data availability information, and you should use
git annex merge
or git annex sync
commands to merge the git-annex branch correctly.
External teaching materials
- git-annex walkthrough from a cognitive scientist (full: 30 min) – a Jupyter notebook; please go through all the items by running the notebook cells or copy/pasting them into a terminal
- git-annex walkthrough (full: 10 min) – original git-annex walkthrough; go through all sections to see which aspects previous walkthroughs did not cover
- (optional) Another walkthrough on a typical use-case for sync’ing)
Exercise: getting data files controlled by git-annex
Using git/git-annex commands
- “Download” a BIDS dataset from https://github.com/datalad/ds000114
getall non-preprocessed T1w anatomicals- Try (and fail) to get all
T1.mgzfiles- Knowing that
yoh@falkor:/srv/datasets.datalad.org/www/workshops/nipype-2017/ds000114is available via http fromhttp://datasets.datalad.org/workshops/nipype-2017/ds000114/.git, get theT1.mgzfilesSolution
% git clone https://github.com/datalad/ds000114 # 1. % cd ds000114 % git annex get sub-\*/anat/sub-\*\_T1w.nii.gz # 2. % git annex get derivatives/freesurfer/sub-\*/mri/T1.mgz # 3. (should fail) % git remote add datalad http://datasets.datalad.org/workshops/nipype-2017/ds000114/.git % git fetch datalad % git annex get derivatives/freesurfer/sub-\*/mri/T1.mgz # 4. (should succeed)
How can we add the file a.txt directly under git, and file b.dat under git-annex?
Simple method (first time)
Use
git addfor adding files under Git, andgit annex addto add files under annex:% git add a.txt % git annex add b.datAdvanced method (for all future
git annex addcalls)If you want to automate such “decision making” based on either file extensions and/or file size, you can specify those rules within a
.gitattributesfile. Adding the following two lines would instruct thegit annex addcommand to add all non-text and all files having the.datextension togit-annexand the rest to git:* annex.largefiles=((mimeencoding=binary)and(largerthan=0)) \*.dat annex.largefiles=anythingNote that the
.gitattributesfile needs to be added and committed in order to come into effect:% git add .gitattributes # to add the new .gitattributes to git % git annex add a.txt b.dat
DataLad
The DataLad project relies on Git and git-annex to establish an integrated data monitoring, management, and distribution environment. As a data distribution capitalizing on a number of “data crawlers” for existing data portals, it provides unified access to over 240TB of data from various initiatives (such as CRCNS, OpenNeuro, etc.).
External teaching materials
- The DataLad Handbook is a code-along crash-course on the basic and advanced principles of DataLad, and the most up-to-date and most comprehensive user-documentation that exists for DataLad. The section Basics (Full: One day) demonstrates and teaches the core commands of the tool, and the section usecases (each usecase: 10-30 min) gives an overview of what is possible.
- DataLad lecture and demo (Full: 55 min) This lecture describes the goals and basic principles of DataLad, and presents the first of the demos on discovery and installation of the datasets.
- DataLad demos of the features (Full: 30 min, review: 10 min) provides an asciinema (and shell script versions) introduction to major features of DataLad.
What DataLad command assists in recording the “effect” of running a command?
% datalad run COMMAND PARAMETERSPlease see datalad run –help for more details.
Exercise: creating, populating, and sharing a new sub-dataset
Using DataLad commands, and starting with your existing clone of
ds000114from the preceding exercise, do the following:
- Create sub-dataset
derivatives/demo-bet- Using a skull-stripping tool (e.g.,
betfrom FSL) to produce a skull-stripped anatomical for each subject under the subdirectoryderivatives/demo-bet; use thedatalad runcommand (available in DataLad 0.9 or later) to keep a record of your analysis- Publish your work to your fork of the repository on GitHub and upload data to your preferred host (an ssh/http server, box.com, Zenodo, Dropbox, etc.)
Solution
% cd ds000114 % datalad create -d . derivatives/demo-bet # 1. % # a somewhat long but fully automated and "protocoled" run solution: % datalad run 'for f in sub-\*/anat/sub-\*\_T1w.nii.gz; do d=$(dirname $f); od=derivatives/demo-bet/$d; mkdir -p $od; bet $f derivatives/demo-bet/$f; done' # 2. % # establish a folder on box.com with access shared among your group % export WEBDAV_USERNAME=secret WEBDAV_PASSWORD=secret % cd derivatives/demo-bet % # see https://git-annex.branchable.com/special_remotes for other supported git-annex special remotes % git annex initremote box.com type=webdav url=https://dav.box.com/dav/team/ds000114--demo-bet chunk=50mb encryption=none % datalad create-sibling-github --publish-depends box.com --access-protocol https ds000114--demo-bet % datalad publish --to github sub\* # 3/ %
Additional relevant helpers
-
sumatra – manages and tracks projects based on numerical simulation or analysis, with the aim of supporting reproducible research; it can be thought of as an “automated electronic lab notebook” for simulation/analysis projects
-
noworkflow – captures a variety of provenance information and provides some analyses, such as graph-based visualization, differencing over provenance trails, and inference queries
-
etckeeper – a helper tool for administering Linux-based systems, which stores and automatically commits any changes in
/etcinto a VCS of your choice; you can track changes in your system configuration, which can be indispensable during system malfunction troubleshooting
Neuroimaging ad-hoc “versioning”
- AFNI captures provenance information in output files that can
be inspected using the
3dinfocommand
Key Points
Using VCS not only improves sharing and collaboration, but is integral for guaranteeing reproducibility
VCS can be used directly or can serve as a foundation for domain-specific tools